- Research
- Open access
- Published:
Multiplex CRISPR/Cas9-mediated genome editing of the FAD2 gene in rice: a model genome editing system for oil palm
Journal of Genetic Engineering and Biotechnology volume 19, Article number: 86 (2021)
Abstract
Background
Genome editing employing the CRISPR/Cas9 system has been widely used and has become a promising tool for plant gene functional studies and crop improvement. However, most of the applied CRISPR/Cas9 systems targeting one locus using a sgRNA resulted in low genome editing efficiency.
Results
Here, we demonstrate the modification of the FAD2 gene in rice using a multiplex sgRNA-CRISPR/Cas9 genome editing system. To test the system’s efficiency for targeting multiple loci in rice, we designed two sgRNAs based on FAD2 gene sequence of the Oryza sativa Japonica rice. We then inserted the validated sgRNAs into a CRISPR/Cas9 basic vector to construct pYLCRISPRCas9PUbi-H:OsFAD2. The vector was then transformed into protoplast cells isolated from rice leaf tissue via PEG-mediated transfection, and rice calli using biolistic transformation. Direct DNA sequencing of PCR products revealed mutations consisting of deletions of the DNA region between the two target sgRNAs.
Conclusion
The results suggested that the application of the multiplex sgRNA-CRISPR/Cas9 genome editing system may be useful for crop improvement in monocot species that are recalcitrant to genetic modification, such as oil palm.
Background
Genetic engineering of crops by the clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein 9 (Cas9) genome editing is the most important breakthrough technology of the decade. CRISPR/Cas9 system adapted from immune system type II, which has been found in bacteria Streptococcus pyogenes. This CRISPR/Cas9 system can efficiently induce targeted mutations based on base-pairing of the engineered single-guide RNAs (sgRNAs) to the target DNA sites [1]. The process relies on the use of sequence-specific nucleases that can be induced to recognize specific DNA sequences and subsequently generate double-strand breaks (DSBs). The DSBs are then repaired by two endogenous mechanisms by either non-homologous end joining, which lead to small insertion or deletion of nucleotides, thereby causing gene knockouts [2], or by homologous recombination, which can cause gene replacements and insertions [3] that lead to the loss of gene function. The CRISPR/Cas9 system is a powerful and efficient tool for targeted mutagenesis with the potential for editing multiple genomic loci and generating a range of gene mutations [4]. There have been several recent reports of CRISPR/Cas9-mediated targeted mutagenesis in a variety of monocots, including rice (Oryza sativa), wheat (Triticum sp.), barley (Hordeum vulgare), and maize (Zea mays) [5,6,7,8,9,10].
CRISPR/Cas9 has also been widely used for functional study of rice genes which was first reported in 2013 with the rice phytoenedesaturase as the target gene [11]. This approach has also been used to develop rice plants with high oleic acid content [12]. High oleic oil is high in monounsaturated fats and low in polyunsaturated fats, characterized at 70% or more oleic acid content. Increasing the monounsaturated fatty acids would enhance the physical properties that can be used in products that need to be shelf-stable with a longer shelf life, a higher heat tolerance, and more nutritional aspects [12]. Besides, a large amount of monounsaturated fat in high oleic oil has been proven to help prevent lifestyle diseases and increase health benefits [13]. Modification of the fatty acid synthesis pathway via suppression of the oleoyl-CoA desaturase (FAD2) will result in the reduction of linoleic acid formation and increase in oleic acid accumulation. In rice genome, four FAD2 genes, designated as OsFAD2-1, OsFAD2-2, OsFAD2-3 and OsFAD2-4 were identified [13]. Among them, the OsFAD2-1 gene is the most highly expressed FAD2 gene in rice seeds. Thus, the knockout of OsFAD2-1 by CRISPR/Cas9 could lead to a high oleic acid content in rice. Previously, a CRISPR/Cas9 vector harbouring single sgRNA targeting OsFAD2-1 gene has been constructed and transformed into rice calli through Agrobacterium-mediated transformation [12]. Rice transgenic mutants were successfully generated, and the impact on fatty acid profiles was evaluated. However, the significant increase of oleic content could only be observed in T2 rice homozygous mutant lines. The oleic acid content in T2 rice homozygous for the OsFAD2-1 mutant allele increased to more than double, from 32 to 80% of the total fatty acid composition, and interestingly, no linoleic acid was detected. Meanwhile, in T2 heterozygous mutant lines, the oleic acid content only increased slightly at about 10% with a concomitant 10% reduction of linoleic acid content as compared with wild-type rice. The study indicated that using a single sgRNA to knockout the OsFAD2-1 gene is not very efficient as homozygous mutant lines could only be obtained in T2 generation lines.
Therefore, in this study, multiple sgRNAs were used to test the plant genome editing system to have a higher homozygous and biallelic mutation rate in T0 plants to get higher oleic acid–producing plants. The findings of previous studies [14,15,16] demonstrated that targeting two or more sgRNAs for one gene were highly efficient and showed high homozygous and biallelic mutations rate in the T0 plants. Studies from [15, 16] resulted in high mutation rates of 82.2%, which subsequently generated 28.1% of the homozygous and 56.7% biallelic mutant lines. The gene mutations were stable to the next generation (T1) following the classic Mendelian law. However, several factors could affect the specificity and efficiency of sgRNAs. Thus, it is recommended that multiple sgRNAs be selected to target a gene of interest.
This study aims to get high oleic acid–producing plants by manipulating the OsFAD2-1 gene in rice but using a multiplex sgRNA-CRISPR/Cas9 genome editing system. We used a robust CRISPR/Cas9 vector system utilizing a plant codon-optimized Cas9 gene as a convenience system [17] to manipulate the OsFAD2-1 gene in rice by employing multiple sgRNAs for high-efficiency genome editing capabilities. Using the vector system, we have successfully constructed a pYLCRISPR/Cas9 plasmid with two different CRISPR/Cas9 target sites within the rice OsFAD2-1 gene, namely OsFAD2-T1 and OsFAD2-T2. We then delivered the plasmid DNA into protoplasts by a PEG-mediated method and at the same time into rice calli by biolistic transformation. We found nearly a 300-bp deletion from both transformation experiments. As rice is a model plant for monocots, the vector system and information obtained from this study can also be used for crops with more complicated genomes, such as oil palm, for genome research of gene function and trait improvement.
Methods
Design of sgRNA
The target sequences of sgRNA were selected and designed using the online tool CRISPR-GE (http://skl.scau.edu.cn/) [17]. The sgRNA sequences were designed based on the genomic sequences of OsFAD2-1 fatty acid desaturase DES2 [O. sativa Japonica Group (Japanese rice)] located on chromosome 2 (NCBI link: https://www.ncbi.nlm.nih.gov/gene/4330523). The CRISPR-GE can detect Protospacer Adjacent Motif (PAM) sequences and lists possible sgRNA sequences within a specific DNA region. A unique genomic potential target of 20 nucleotide DNA sequence present immediately upstream of a PAM site at 3′ (5′-20nt-NGG-3′) was selected by comparing to the rest of the genome. Potential target sequences with dozens of bases forward and reverse were examined for sequence similarity using Blast at NCBI (https://www.ncbi.nlm.nih.gov) against the rice (O. sativa japonica) genome with somewhat similar sequences option. This was carried out to confirm their targeting specificity in the genomes. Two candidates of target sgRNA sequences were selected based on their GC content (greater than 40% for high editing efficiency), and the absence of four or more consecutive “T” nucleotides in the target sequence as the sequence would be recognized as a transcriptional termination signal by RNA polymerase III [16]. The secondary structures of target-sgRNA sequences (20-bp target) linked to nucleotides of the sgRNA scaffold (GTTTTAGAGCTAGAAATAGCAAGTTAAAATAAGGCTAGTCCGTTATCAACTTGAAAAAGTGGCACCGAGTCGGTGCTTTTTTT) were analysed with the RNA Folding Form programme (http://mfold.rna.albany.edu/?q=mfold/RNA-Folding-Form2.3). The software was used to ensure good base pairings between the target sequence and the sgRNA sequence, greatly affecting the editing efficiency [18].
In vitro validation of sgRNA
An in vitro assay to determine the most effective sgRNAs was carried out prior to delivery into the cells. The efficiencies of sgRNAs were evaluated by using the Guide-it™ sgRNA In Vitro Transcription and Screening Systems Kit (Takara Bio USA, Inc). Briefly, PCR was performed with a 56- to 58-nt customized oligonucleotide forward PCR primer (Supplementary Table 1) that contained the T7 promoter (TAATACGACTCACTATA), the 20-nt sgRNA DNA targeting sequence, and the 15-nt scaffold template annealing (GTTTAAGAGCTATGC). PCR amplification was performed using the following condition: 33 cycles of 98 °C for 10 s and then 68 °C for 10 s. PCR product of ~ 130 bp was purified and used as a template for a T7 RNA polymerase–mediated transcription reaction using Guide-it T7 Polymerase Mix (Takara Bio USA, Inc). The transcribed sgRNAs were then purified, and the concentration was determined using a NanoDrop 2000 spectrophotometer (Thermo Fisher). The transcribed sgRNA, Cas9 protein, and targeted DNA fragment were mixed for in vitro cleavage assay according to the manufacturer’s protocol (Takara Bio USA, Inc). The resulted cleavage assay products were electrophoresed on a 2% agarose gel to determine the cleavage potential at the DNA target site. Trans2K® Plus II DNA Marker (Cat. No. BM111-01, TransGen Biotech, China) and 100 bp plus DNA Ladder (Cat. No. BM321-01, TransGen Biotech, China) were used as molecular weight standards for agarose gel electrophoresis.
Construction of multiplex sgRNA CRISPR/Cas9 vector
In order to construct the CRISPR/Cas9 vector with multiplex sgRNA, two intermediate expression cassettes containing the sgRNA sequence were constructed according to the method described by Ma et al. [17]. Briefly, the 20-nt target sequences were synthesized using overlapping PCR. In the first round of PCR, the 20-nt target sequences were ligated into its corresponding sgRNA expression cassettes (pYLsgRNA-OsU6a and pYLsgRNA-OsU6b, obtained from Dr. Yao-Guang Liu’s laboratory, South China Agricultural University, China). This was followed by a second PCR to combine the two fragments of sgRNA expression cassettes flanked with two BsaI restriction sites. Amplified PCR products containing sgRNA expression cassettes were then cloned into BsaI site of pYLCRISPR/Cas9PubiH (obtained from Dr. Yao-Guang Liu’s laboratory, South China Agricultural University, China), via the Gibson assembly cloning method [19]. Positive clones were confirmed by PCR, AscI digestion, and DNA sequencing. All the primer sequences used in this study are listed in Supplementary Table 1.
Plant materials
Mature seeds of rice (O. sativa L.) cultivar Nipponbare were dehulled and sterilized with 70% ethanol for 1 min (Supplementary Fig. 1). The seeds were further sterilized with 2.5% sodium hypochlorite for 20 min and washed at least five times with sterile water. The sterile seeds were then incubated on 1/2 MS medium with a photoperiod of 12-h light and 12-h dark at 26 °C for 10–14 days. Two-week-old plants were used for protoplast isolation. Meanwhile, rice calli (O. sativa L.) cultivar Nipponbare were cultured on a callus induction medium for 1 month. Embryogenic calli obtained were used as target material for bombardment.
Protoplast isolation and PEG-mediated protoplast transfection
Rice protoplasts were isolated following the methods described by Zhang et al. [20] and Yoo et al. [21] with some modifications. The green tissues from the stem and sheath of two-week-old rice seedlings were used for protoplast isolation (Fig. 1A). About 30 seedlings were cut into 0.5-mm strips with a sterile razor blade (Fig. 1B). The leaf strips were immediately immersed into 0.6 M mannitol for 10 min in the dark (Fig. 1C). The strips were then incubated in an enzyme solution which consisted of 1.5% (w/v) cellulase R10, 0.75% (w/v) macerozyme R-10, 0.6 M mannitol, 10 mM MES at pH 5.7, 10 mM CaCl2, and 0.1% (w/v) BSA for 6 h in the dark with gentle shaking (60–80 rpm). An equal volume of W5 solution (154 mM NaCl, 125 mM CaCl2, 5 mM KCl, and 2 mM MES at pH 5.7) was added, and the digestion mixture was gently shaken for 1 h. The released protoplasts were filtered through a 40-μm nylon mesh into a round bottom tube. The protoplasts were collected by centrifugation at 1000 rpm for 5 min with a swing bucket. After one washing step with W5 solution, the pelleted protoplasts were resuspended in 500 ml MMG solution (0.4 M mannitol, 15 mM MgCl2, and 4 mM MES at pH 5.7). The protoplasts were transferred to a slide, and the viability was checked by microscopy (Olympus, Tokyo, Japan) (Fig. 1D). For each transfection, 2 μg of total plasmid DNA were mixed with 100 μl protoplasts. Then, 100 μl freshly prepared PEG solution [40% (w/v) PEG 4000, 0.2 M mannitol, and 0.1 M CaCl2] was added, and the mixture was gently mixed and incubated at room temperature for 20 min in the dark. After incubation, 1 ml W5 solution was added. The resulting solution was mixed well by gently inverting the tube, and the protoplasts were pelleted by centrifugation at 1000 rpm for 5 min. The protoplasts were resuspended gently in 1 ml W5 solution. Finally, the transfected protoplasts were incubated in the dark at room temperature for 24–48 h.
Genomic DNA extraction and mutation validation in transfected protoplasts
Genomic DNA was extracted from pooled protoplasts with four replications. The protoplasts cells were lysed by adding an equal volume of chloroform and allowing the phases to separate by centrifugation at 13,000 rpm for 15 min. The clear upper layer (aqueous phase) was transferred into a new tube, and an equal volume of isopropanol was added. The mixture was mixed gently and incubated at – 80 °C. After 1 h, the DNA was collected by centrifugation at 13,000 rpm for 15 min. The supernatant was discarded, and the pellet was washed with 1 ml 100% ethanol. The ethanol was discarded, and the pellet was air-dried and dissolved in 50 μl sterile water. Two pairs of primers were designed to amplify the targeted genomic region. The 25 μl PCR reaction contained approximately 100 ng template DNA, 1 μl (10 μM) specific forward and reverse primers, and 12.5 μl of 2× T5 Super PCR Mix (TsingKe Co. Ltd., Beijing, China). The primers used are listed in Supplementary Table 1. The purified PCR products were sequenced. Targeted gene mutations were detected by aligning the sequencing chromatograms of these PCR products with the wild-type controls using the SNAPGENE Tools software.
Biolistic transformation of rice calli
Plasmid DNA for the bombardment of rice calli was isolated using a QIAGEN Hi-Speed Plasmid Midi Kit (Qiagen, USA) according to the manufacturer’s instructions. A total of 5 μg/μl of plasmid DNA was mixed with 0.6-μm gold particles. About 30 to 40 embryogenic calli with 3- to 4-mm sizes were arranged at the centre of the plate containing a callus initiation medium. The calli were bombarded at 1100 psi, with a target distance of 60 mm, using the PDS-1000/He™ device according to manufacturer instruction (Bio-Rad, USA). The bombarded calli were incubated in the dark at 26 °C for callus initiation.
Genomic DNA extraction and mutation detection in bombarded calli
Total genomic DNA of bombarded rice calli (1 to 5 mg) was isolated 2 months after transformation according to CTAB protocol [22]. The extracted genomic DNA was then used as a template to amplify the desired fragments in each of the target genes using primers flanking the target site (Supplementary Table 1). PCR amplification reaction was performed using KOD-FX DNA Polymerase (TOYOBO-KOD FX) in a final volume of 20 μl. PCR was carried out using an Eppendorf AG 22331 Hamburg Mastercycler (Eppendorf) for 36 cycles consisting of denaturation at 95 °C for 30 s, annealing at 60 °C for 30 s, and extension at 72 °C for 30 s. The amplified products were cloned into the pEASY-Blunt cloning vector (TransGen Biotech, China). The resulting ligation product was subsequently transformed into Escherichia coli Trans1-T1 competent cell and spread onto LB plates supplemented with kanamycin. Putative colonies were screened by a colony PCR, and the insert DNA was sequenced. The mutations were analysed by DSDecodeM (http://skl.scau.edu.cn/dsdecode/), an online analysis tool [17] for decoding multiple Sanger sequencing chromatograms derived from PCR amplicons. The degenerate sequence decodes superimposed sequencing peaks containing various types of mutations such as biallelic, heterozygous, and homozygous mutations. The sequences (ab1 format) with the sequence information and the reference sequence (WT) were uploaded and decoded. The results were to verify the deletions, insertions, substitutions, and type of mutations in the target region of sgRNA.
Results
sgRNA design and validation of cleavage efficiency via in vitro Cas9 cleavage assay
In order to disrupt the FAD2 gene, two target sites on its coding sequence (CDS) region located on the exon two were selected, and the primer pairs (OsFAD2#220F and OsFAD2#1136R) flanking the two sgRNAs were designed (Fig. 2A). The two designed sgRNAs, namely OsFAD2-T1 (5′ TACGTGTACCACAACCCGATCGG 3′) and OsFAD2-T2 (5′ CTACCTGCAGCACACCCACCCGG 3′), comprised of 20 nucleotide sequences and immediately followed by nucleotides of the PAM sequences (5′-NGG). The selected sgRNAs had 50% and 65% GC content, respectively. For the selection of efficient sgRNAs that affect the editing ability of the CRISPR/Cas9 system, sgRNAs with a GC content between 50 and 75% are desirable [16]. This range of GC contents in the target sequences could result in relatively higher editing efficiencies [17].
A Schematic of OsFAD2-1 gene loci. Predicted secondary structures for (B) sgRNA1 and (C) sgRNA2. PCR amplification and validation of sgRNAs via in vitro Cas9 cleavage assay; D PCR product from amplification of sgRNAs template at ~ 130 bp (lane 1: sgRNA1, lane 2: sgRNA2) for in vitro transcription, E PCR amplification of rice target DNA region (OsFAD2T1T2) at ~ 917 bp, and (F) analysis of cleavage products for sgRNA1 at 312 bp and 582 bp (lane 1) and sgRNA2 at 614 bp and 280 bp (lane 2). Band size at ~ 130 bp in (F) at lanes 1 and 2 are the residue of sgRNA template. Red arrows indicate the band size at the expected molecular weight. Lane M is Trans2K® Plus II DNA Marker (TransGen Biotech, China)
Based on the secondary structure of target sequences, the predicted number of unpaired or mismatched nucleotides in the targeted regions of the RNA sequence for both sgRNAs, OsFAD2-T1 and OsFAD2-T2, was shown to contain no more than six mismatches to the target sequences (Fig. 2). The two structures were predicted for sgRNA1 (OsFAD2-T1) containing 4 and 5 mismatched nucleotides (Fig. 2B) and three structures containing 0, 3, and 6 mismatches for sgRNA2 (OsFAD2-T2) (Fig. 2C). The predicted secondary structure of the OsFAD2-T1 and OsFAD2-T2 seed regions may potentially improve the cleavage activity of Cas9 and sgRNA performance. It was suggested that the secondary structure of sgRNA is an important parameter and critical factor to achieve high on-target efficiency of the CRISPR/Cas9 system [23].
A fast and sensitive in vitro Cas9 cleavage assay for sgRNA validation was also done before applying the sgRNAs into plant cells. The main advantage of in vitro Cas9 cleavage assay is that the evaluation of the target specificity in vitro is independent of the genome or species used. As shown in Fig. 2D, 130-bp PCR templates for in vitro transcription of sgRNAs were successfully amplified. The PCR products containing the T7 promoter sequence fused with the variable 20-nt sgRNA target sequence (OsFAD2-T1 and OsFAD2-T2) followed by constant region sequences. The PCR products were then transcribed in vitro with T7 polymerase–synthesized sgRNAs. A 917-bp PCR amplicon containing a target region was amplified using primers, OsFAD2#220F and OsFAD2#1136R, from wild-type Japonica rice cultivar Nipponbare genomic DNA (Fig. 2E). According to the protocol, the targeted DNA fragment, sgRNAs, and recombinant Cas9 nuclease were combined for in vitro cleavage reaction. The efficiency of two sgRNAs to cleave the target DNA fragment was verified when no band at a size of 917 bp was observed (Fig. 2F). The 917-bp DNA fragment was completely cleaved by sgRNA1/Cas9 ribonucleoprotein complexes to form two DNA fragments with sizes of 312 bp and 582 bp. Meanwhile, two fragments of sizes 614 bp and 280 bp were observed from sgRNA2/Cas9 ribonucleoprotein complexes, suggesting both sgRNAs were efficient to introduce mutation or deletion within the target region.
CRISPR/Cas9 expression vector construction
In order to test the ability and efficiency of multiple sgRNA for genome editing in rice, two sgRNAs expression cassettes targeting two adjacent sites of the OsFAD2-1 gene were assembled in a single vector. These two sgRNA sequences are driven by their own individual promoter, which has been constructed using a robust CRISPR/Cas9 vector system developed by Ma et al. [17]. As illustrated in Fig. 3, two multiple sgRNA expression cassettes were assembled into the BsaI-linearized pYLCRISPR/Cas9Pubi-H binary vector by the Gibson assembly method to create pLYCRISPRCas9PUbi-H:OsFAD2. The E. coli colonies carrying the pLYCRISPRCas9PUbi-H:OsFAD2 plasmid were identified by colony PCR and restriction enzyme digestion with Asc1 (Supplementary Fig. 2). In addition to these molecular detection methods, validation by Sanger sequencing of the positive colonies using the primer pair SP-L1 and SP-R was also performed. Nucleotide blast from the NCBI webpage used for sequence comparison confirmed that the PCR product contained the vector sequences (supplementary Fig. 3).
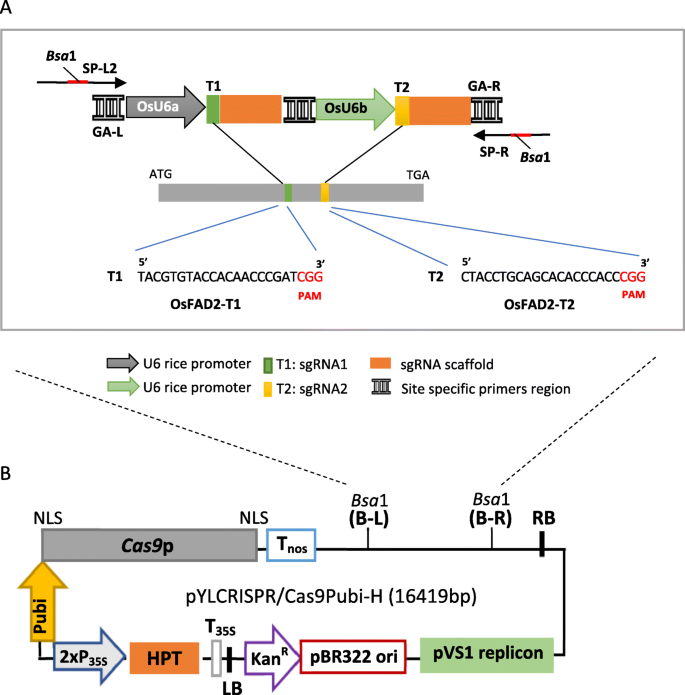
Strategy for the construction of pYLCRISPR/Cas9 targeting OsFAD2-1 in rice. A Schematic illustration of target sites for two sgRNA expression cassettes driven by U6 promoters from rice; OsU6a promoter for OsFAD2-T1 and OsU6b promoter for OsFAD2-T2, the 20-nt sequences in green square boxes and orange square boxes indicate the PAM motif of each sgRNA, the BsaI restriction enzyme sites for the entry of multiple sgRNA expression cassettes, SP-L2 and SP-R are sequencing vector-specific primers, GA-L and GA-R are GA site-specific primers for the Gibson assembly of sgRNA expression cassettes; B pYLCRISPR/Cas9PubiH basic vector has two BsaI sites for insertion of two sgRNA expression cassettes [17] (LB: left border of T-DNA, T35S: cauliflower mosaic virus 35S terminator, HPT: gene for hygromycin phosphotransferase, 2xP35S: Double cauliflower mosaic virus 35S promoter, Pubi: maize ubiquitin Ubi1 promoter, NLS: Nuclear localization sequence. Cas9p: CRISPR associated protein 9, Nos: nopaline synthase gene terminator, RB: right border of T-DNA, pVS1 replicon: pVS1 for replication, pBR322: pBR322 vector, KanR: Kanamycin resistance gene)
The pYLCRISPRCas9PUbi-H:OsFAD2 vector utilizing a plant codon-optimizedCas9 gene (Cas9p) was used to induce and improve the editing efficiency in monocots using rice as a testing platform [17]. In addition, the vector also contains an RNA polymerase III (Pol III) promoter, OsU6, a monocot-derived promoter, as a strategy to increase the transcription of the sgRNA in monocot plants. It was reported that sgRNA regulated by the OsU6 promoter produced more transcript levels than the OsU3 promoter [24]. Due to this, pYLCRISPRCas9PUbi-H:OsFAD2 has two types of OsU6 promoters, namely OsU6a and OsU6b. The OsU6a is derived from an Indica rice cultivar 93-11, while OsU6b originated from a Japonica rice cultivar Nipponbare [17]. Both promoters are used as a strategy to control multiple sgRNA. The pYLCRISPRCas9PUbi-H:OsFAD2 binary vector also contains the hygromycin phosphotransferase (HPT) gene driven by the maize ubiquitin promoter for the selection of transgenic plants.
Detection of multiplex genome editing in transfected protoplasts and bombarded calli
PEG-mediated transfection of employing rice protoplasts isolated from young green tissue is an alternative testing strategy before performing a stable transformation. This is an easy and fast procedure to evaluate the CRISPR/Cas9 genome editing system. Several studies have shown the use of protoplasts as a useful and convenient protocol to determine the effectiveness of designed sgRNAs and confirm the sgRNA/Cas9 editing efficiency [11, 15, 25,26,27,28,29,30,31].
The transfected protoplasts with four replications were isolated for genomic DNA isolation. The genomic DNA was used as a PCR template for the target gene amplification (OsFAD2-T1 and OsFAD2-T2). Two sets of primers were used for their specificities and sensitivities in PCR for OsFAD2-1 gene detection. The first pair, OsFAD2-F and OsFAD2-R primers generate two fragments of 502 bp and 200 bp (Fig. 4A). Meanwhile, the second set of primers (OsFAD2-R and OsFAD2-F2) generated amplicons with sizes of 776 bp and 474 bp (Fig. 4B). However, the amplification with the second set of primers seems to be more suitable for high-quality sequencing analysis of about 400 to 700 bp [32]. Double bands obtained could be due to the indel mutation generated. PCR products obtained using OsFAD2-R and OsFAD2-F2 primers flanking the targeted region at 474 bp were directly sequenced.
PCR-amplified DNA from protoplasts transfected with pYLCRISPRCas9Pubi-H:OsFAD2. A DNA fragments of 502 bp and 200 bp were amplified using FAD2-F and FAD2-R primers. B DNA fragments of 776 bp and 474 bp were amplified from second set PCR using FAD2-F2 and FAD2-R primers. Lane M is Trans2K® Plus II DNA Marker (TransGen Biotech, China), and lanes marked with “+” are positive samples. C PCR products at 474 bp were directly sequenced and analysed using SNAPGENE revealed mutations consisting of deletions in the DNA region between sgRNA1and sgRNA2. The blue background part is the deleted sequences; the first line (sequence of the wild type target gene) and target site in grey boxes were highlighted. D Sequencing chromatogram, the red box is the residue of target 1 (OsFAD2-T1), and the green box is the residue of target 2 (OsFAD2-T2)
The sequences were aligned with the reference sequence of the OsFAD2-1 and compared to the rice genome. Two (events 1 and 2) out of 4 events tested showed deletions of 302 bp, suggesting that Cas9 cuts 3 bp upstream of the PAM for sgRNA1 and 3 bp upstream of the PAM for sgRNA2 (Fig. 4C). The sequencing chromatogram consisting of the residues of sgRNA1 and sgRNA2 after the deletion occurred is shown in Fig. 4D. This observation demonstrated that the non-homologous end-joining repair mechanism is used to repair the DSBs created by the Cas9. Previous studies in monocot also reported that using multiple sgRNA target regions resulted in targeted deletions of 10-bp to over 200-kb nucleotides between two target sites in maize [33].
On the other hand, the biolistic-mediated method was used to generate transgenic rice lines transformed with the plasmid DNA, pYLCRISPRCas9PUbi-H:OsFAD2. Two weeks after the bombardment, the bombarded rice calli were cultured on hygromycin selection media for 2 months. Hygromycin-resistant calli produced were then subjected to PCR and DNA sequencing to detect mutations (Supplementary Fig. 4). Twenty-four resistant calli (T0) were randomly selected for PCR analysis. Of these, five samples showed amplified PCR products at 776 bp and 474 bp were directly sequenced. Sequence analysis of PCR amplicon confirmed the mutant status. The deletion was between sgRNA1 and sgRNA2 target sites, precisely occurred as expected (Supplementary Fig. 4B and 4C). The sequencing result of amplicon derived from transformants was also decoded using DSDecodeM [32] (Supplementary Fig. 5). Two different homozygous mutant alleles for two amplified PCR products were detected. A nucleotide (1 bp) insertion for amplicon 776 bp and a large deletion of 302 nucleotides for smaller amplicon 474 bp in both alleles were identified in the T0 rice. Even though there was no result from fatty acid analysis from this study, the detection of homozygous mutations through molecular analyses probably could result in the knockout of the OsFAD2-1 gene, subsequently increasing the oleic acid content in rice seeds. This result is consistent with a previous study that reported homozygous mutants in the rice T0 [34].
Discussion
The development and improvement of the CRISPR/Cas9 system are being established as the preferred method for gene manipulation in the crop. Its efficiency has been demonstrated in model plants such as Arabidopsis, rice, and tobacco [35,36,37]. Rice is a diploid and monocot with a small genome size (430 Mb) plant that has been one of the choices for CRISPR/Cas9 application model systems. Furthermore, DNA transformation of rice is efficient and relatively easy to produce transgenic plants, making it a useful plant for studying biology. Therefore, in this study, we used rice as a model system in the attempt to develop a genome editing system for oil palm. The genomic sequences of the rice (O. sativa) genome were found to have significant similarities to the oil palm genomic sequences [38, 39], which can be exploited in oil palm research. This also provides an opportunity to further investigate molecular genetics linked to other oil palm traits by using rice as the reference genome.
Thus, in parallel to this research, efforts are focused on achieving the ultimate goal of the oil palm genetic engineering programme to increase the production of oleic acid content in palm oil by manipulating the FAD2 gene of the fatty acid biosynthesis pathway [40,41,42]. In plant, reducing or knocking out the expression of the FAD2 gene will reduce the percentage of linoleic acid and subsequently will increase the level of oleic acid. Most plants have several FAD2 genes; rice has four FAD2 genes and oil palm has two FAD2 genes, namely EgFAD2-1 and EgFAD2-2. In this work, a robust CRISPR/Cas9 system for multiplexed plant genome editing originally developed by Ma et al. [17] has been conveniently utilized to target the FAD2 gene of the rice genome. This system is efficient for linking multiple sgRNA cassettes in a single reaction. More than one sgRNA was recommended to target multiple sites in a single gene to improve editing rates, especially in crops with low transformation efficiencies such as oil palm. Currently, the oil palm transformation efficiency is 0.7% for Agrobacterium-mediated plant transformation [43], 1.5% for particle bombardment method [44], and 14% for protoplast DNA microinjection [45].
To further improve the transformation efficiency, the potential of the multiplex CRISPR/Cas9 technique was explored by designing two sgRNAs, OsFAD2-T1 and OsFAD2-T2, to interrupt the rice OsFAD2-1 gene function. For efficient sgRNA target selection, highly specific nucleotide sequences to the genome with GC content 50% and 65% were preferred to reduce off-target effects and produce high editing efficiency. In addition, a plant codon-optimized Cas9 gene (Cas9p) with higher GC contents (62.5%) in the 5′ region (400 bp) has been used to direct the sgRNAs efficiently. The multiplex CRISPR/Cas9 vector cassettes were created with a construct composed of Cas9 expressed under the control of Pol II Ubi1 promoter. Meanwhile, the Pol III OsU6 (OsU6a and OsU6b) promoter was used to drive the expression of sgRNA for higher genome editing efficiency. Previous reports in monocots showed that Cas9 expression driven by a strong constitutive promoter maize ubiquitin (Ubi1) and the rice U6 (OsU6) promoter for sgRNA expression is relatively efficient in generating targeted effects in the transformed plants (T0) [26, 46, 47].
A pYLCRISPRCas9PUbi-H:OsFAD2 vector with two sgRNA expression cassettes was efficiently assembled in one round of cloning by the Gibson assembly. The integration of the CRISPR/Cas9 cassettes was confirmed by PCR and Sanger sequencing. The vector was tested through the PEG-protoplast-mediated transformation and was then successfully used to create mutations in the stable transgenesis by using a biolistic transformation system. The transgenic lines generated were randomly picked and further genotyped by genomic PCR and followed by direct Sanger sequencing to determine the mutation efficiency. Knocking out the two sgRNAs targeting a single gene caused large deletion of DNA fragments between two target sites and subsequently generated homozygous mutant allele T0 rice calli. This result is also consistent with a previous study which reported the generation of homozygous mutants in the rice T0 [34]. Rice plants with high oleic acid have been developed by other research groups using different approaches, including RNAi [13] and CRISPR/Cas9 single sgRNA gene editing [12]. The oleic acid content was increased to 51.7% as compared to wild type ~ 32% via RNAi technology and increased to approximately 80% via the CRISPR/Cas9 experiment.
Numerous reports of CRISPR/Cas9 targeting of the FAD2 gene have been published including in Camelina sativa [48, 49], soybean (Glycine max) [50, 51], Brassica napus [52], peanut [53], and tobacco [54]. Results indicated that mutations were obtained at the specific sites in the FAD2 gene at any location by gene knockout. Moreover, the mutation efficiency mediated by the CRISPR/ Cas9 system varies in different plant species [36, 14]. This finding augurs the finding by Doudna et al. [55], which indicated that the CRISPR/Cas9 genome editing strategy is a way to precisely edit and modify any region of the genome in any species. Later, this technology will be applied to oil palm with the aim to produce high oleic acid content. For this purpose, we aim to knock out the activities of the two key enzymes regulating oil palm fatty acid composition, namely the FAD2 and palmitoyl-ACP- thioesterase (PAT) genes [40,41,42]. This strategy would greatly reduce the FAD2 and PAT enzyme activities and is expected to increase the oleic acid content to more than 70% with a concomitant decrease of linoleic acid and other long-chain polyunsaturated fatty acids. A high expression of Cas9 and sgRNA driven by plant endogenous promoters is essential for developing the CRISPR/Cas9 system in plants. The Pol III promoter (U6) of oil palm will be used to control the expression of sgRNA since the use of endogenous U6P promoter increased the gene-editing efficiency 4–6-fold as demonstrated in soybean [50] and rice [17]. The oil palm constitutive promoters (UEP1, UEP2, TCTP), which were isolated and characterized previously [41], will also be incorporated in the CRISPR/Cas9 vector to control the Cas9 gene. The strategies are expected to increase the gene-editing efficiency of oil palm significantly.
Conclusion
CRISPR/Cas9 genome editing technology was tested in rice as a model monocot plant for oil palm using a robust CRISPR/Cas9 system utilizing plant codon-optimized Cas9 gene. Two sgRNAs were designed to knock out the OsFAD2-1 gene in rice using both PEG-mediated transfection and particle bombardment. Large deletions of approximately 300 bp were detected by PCR and further confirmed by Sanger sequencing in both transformed rice protoplasts and rice calli. A homozygous mutation identified in the T0 generation in rice suggested that the multiplex CRISPR/Cas9 system is highly active in rice, which may be related to the targeting efficiency of different sgRNAs. This study suggested a robust CRISPR/Cas9 vector system could become a powerful tool and can be utilized for efficient multiplex genome editing in future applications. The successful application of genome editing using the CRISPR/Cas9 approach may provide effective solutions for the more complicated crop. This study paves the way for future research in developing oil palm to produce higher oleic acid via Cas9/sgRNA-mediated knockouts of the fatty acid desaturase genes.
Availability of data and materials
The authors declare that all data generated or analysed in this study are included in the article.
Abbreviations
- Cas9:
-
CRISPR-associated protein 9
- CRISPR:
-
Clustered regularly interspaced short palindromic repeats
- FAD2:
-
Oleoyl-CoA desaturase
- OsFAD2:
-
Oryza sativa FAD2
- PAM:
-
Protospacer adjacent motif
- PEG:
-
Polyethylene glycol
- sgRNA:
-
Single-guide RNA
References
Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E (2012) A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337(6096):816–821. https://doi.org/10.1126/science.1225829
Malzahn A, Lowder L, Qi Y (2017) Plant genome editing with TALEN and CRISPR. Cell Biosci 7(1):21. https://doi.org/10.1186/s13578-017-0148-4
Symington LS, Gautier J (2011) Double-strand break end resection and repair pathway choice. Annu Rev Genet 45:247–271. https://doi.org/10.1146/annurev-genet-110410-132435 PMID: 21910633
Komor AC, Badran AH, Liu DR (2017) CRISPR-based technologies for the manipulation of eukaryotic genomes. Cell. 168(1-2):20–36. https://doi.org/10.1016/j.cell.2016.10.044
Wang M, Mao Y, Lu Y, Tao X, Zhu JK (2017) Multiplex gene editing in rice using the CRISPR-Cpf1 system. Mol Plant 10(7):1011–1013. https://doi.org/10.1016/j.molp.2017.03.001
Hu X, Meng X, Liu Q, Li J, Wang K (2018) Increasing the efficiency of CRISPR-Cas9-VQR precise genome editing in rice. Plant Biotechnol J 16(1):292–297. https://doi.org/10.1111/pbi.12771
Zhang Y, Liang Z, Zong Y, Wang Y, Liu J, Chen K, Qiu JL, Gao C (2016) Efficient and transgene-free genome editing in wheat through transient expression of CRISPR/Cas9 DNA or RNA. Nat Commun 7(1):12617. https://doi.org/10.1038/ncomms12617
Gasparis S, Kala M, Przyborowski M, Lyznik LA, Orczyk W, Nadolska-Orczyk A (2018) A simple and efficient CRISPR/Cas9 platform for induction of single and multiple, heritable mutations in barley (Hordeum vulgare L.). Plant Methods 14(1):111. https://doi.org/10.1186/s13007-018-0382-8
Kis A, Hamar E, Tholt G, Ban R, Havelda Z (2019) Creating highly efficient resistance against wheat dwarf virus in barley by employing CRISPR/Cas9 system. Plant Biotechnol J 17(6):1004–1006. https://doi.org/10.1111/pbi.13077
Feng C, Su H, Bai H, Wang R, Liu Y, Guo X, Liu C, Zhang J, Yuan J, Birchler JA, Han F (2018) High-efficiency genome editing using a dmc1 promoter-controlled CRISPR/Cas9 system in maize. Plant Biotechnol J 16(11):1848–1857. https://doi.org/10.1111/pbi.12920
Shan Q, Wang Y, Li J, Zhang Y, Chen K, Liang Z (2013) Targeted genome modification of crop plants using a CRISPR-Cas system. Nat Biotechnol 31(8):686–688. https://doi.org/10.1038/nbt.2650
Abe K, Araki E, Suzuki Y, Toki S, Saika H (2018) Production of high oleic/low linoleic rice by genome editing. Plant Physiol Biochem 131:58–62. https://doi.org/10.1016/j.plaphy.2018.04.033
Zaplin ES, Liu Q, Li Z, Butardo VM, Blanchard CL, Rahman S (2013) Production of high oleic rice grains by suppressing the expression of the OsFAD2-1 gene. Funct Plant Biol 40(10):996–1004. https://doi.org/10.1071/FP12301
Zhang H, Zhang J, Wei P, Zhang B, Gou F, Feng Z, Mao Y, Yang L, Zhang H, Xu N, Zhu J (2014) The CRISPR/Cas9 system produces specific and homozygous targeted gene editing in rice in one generation. Plant Biotechnol J 12(6):797–807. https://doi.org/10.1111/pbi.12200
Xie K, Minkenberg B, Yang Y (2015) Boosting CRISPR/Cas9 multiplex editing capability with the endogenous tRNA-processing system. Proc Natl Acad Sci U S A 112(11):3570–3575. https://doi.org/10.1073/pnas.1420294112
Liang G, Zhang H, Lou D, Yu D (2016) Selection of highly efficient sgRNAs for CRISPR/Cas9 based plant genome editing. Sci Rep 6(1):21451. https://doi.org/10.1038/srep21451
Ma X, Zhang Q, Zhu Q, Liu W, Chen Y, Qiu R, Wang B (2015) A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Mol Plant 8(8):1274–1284. https://doi.org/10.1016/j.molp.2015.04.007
Zuker M (2003) Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 31(13):3406–3434. https://doi.org/10.1093/nar/gkg595
Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CR, Smith HO (2009) Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods 6(5):343–345. https://doi.org/10.1038/nmeth.1318
Zhang Y, Su J, Duan S, Ao Y, Dai J, Liu J, Wang P, Li Y, Liu B, Feng D, Wang J, Wang H (2011) A highly efficient rice green tissue protoplast system for transient gene expression and studying light/chloroplast-related processes. Plant Methods 7(1):30. https://doi.org/10.1186/1746-4811-7-30
Yoo SD, Cho YH, Sheen J (2007) Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc 2(7):1565–1572. https://doi.org/10.1038/nprot.2007.199
Doyle JJ, Doyle JL (1990) Isolation of plant DNA from fresh tissue. Focus. 12:13
Liu X, Homma A, Sayadi J, Yang S, Ohashi J, Takumi T (2016) Sequence features associated with the cleavage efficiency of CRISPR/Cas9 system. Sci Rep 6(1):19675. https://doi.org/10.1038/srep19675
Mikami M, Toki S, Endo M (2015) Comparison of CRISPR/Cas9 expression constructs for efficient targeted mutagenesis in rice. Plant Mol Biol 88(6):561–572. https://doi.org/10.1007/s11103-015-0342-x
Jiang W, Zhou H, Bi H, Fromm M, Yang B, Weeks DP (2013) Demonstration of CRISPR/Cas9/sgRNA-mediated targeted gene modification in Arabidopsis, tobacco, sorghum and rice. Nucleic Acids Res 41(20):e188. https://doi.org/10.1093/nar/gkt780 PMID: 23999092
Xie K, Yang Y (2013) RNA-guided genome editing in plants using a CRISPR-Cas system. Mol Plant 6(6):1975–1983. https://doi.org/10.1093/mp/sst119
Shan Q, Wang Y, Li J, Gao C (2014) Genome editing in rice and wheat using the CRISPR/Cas system. Nat Protoc 9(10):2395–2410. https://doi.org/10.1038/nprot.2014.157 PMID: 25232936
Lowder LG, Zhang D, Baltes NJ, Paul JW, Tang X, Zheng X (2015) A CRISPR/Cas9 toolbox for multiplexed plant genome editing and transcriptional regulation. Plant Physiol 169(2):971–985. https://doi.org/10.1104/pp.15.00636
Woo JW, Kim J, Kwon SI, Corvalan C, Cho SW, Kim H, Kim SG (2015) DNA-free genome editing in plants with preassembled CRISPR-Cas9 ribonucleoproteins. Nat Biotechnol 33(11):1162–1164. https://doi.org/10.1038/nbt.3389
Li QL, Zhang DB, Chen MJ, Liang WQ, Wei JJ, Qi YP, Yuan Z (2016) Development of japonica photo-sensitive genic male sterile rice lines by editing carbon starved anther using CRISPR/Cas9. J Genet Genomics 43(6):415–419. https://doi.org/10.1016/j.jgg.2016.04.011
Wang F, Wang C, Liu P, Lei C, Hao W, Gao Y, Liu YG (2016) Enhanced rice blast resistance by CRISPR/Cas9-targeted mutagenesis of the ERF transcription factor gene OsERF922. PLoS One 11(4):e0154027. https://doi.org/10.1371/journal.pone.0154027
Liu W, Xie X, Ma X, Li J, Chen J, Liu YG (2015) DSDecode: a web-based tool for decoding of sequencing chromatograms for genotyping of targeted mutations. Mol Plant 8(9):1431–1433. https://doi.org/10.1016/j.molp.2015.05.009
Xing HL, Dong L, Wang ZP, Zhang HY, Han CY, Liu B (2014) A CRISPR/Cas9 toolkit for multiplex genome editing in plants. BMC Plant Biol 14(1):327. https://doi.org/10.1186/s12870-014-0327-y
Liao S, Qin X, Luo L, Han Y, Wang X, Usman B, Nawaz G, Zhao N, Liu Y, Li R (2019) CRISPR/Cas9-induced mutagenesis of semi-rolled Leaf1, 2 confers curled leaf phenotype and drought tolerance by influencing protein expression patterns and ROS scavenging in Rice (Oryza sativa L.). Agronomy 9(11):728. https://doi.org/10.3390/agronomy9110728
Belhaj K, Chaparro-Garcia A, Kamoun S, Nekrasov V (2013) Plant genome editing made easy: targeted mutagenesis in model and crop plants using the CRISPR/Cas system. Plant Methods 9(1):39. https://doi.org/10.1186/1746-4811-9-39
Fauser F, Schiml S, Puchta H (2014) Both CRISPR/Cas-based nucleases and nickases can be used efficiently for genome engineering in Arabidopsis thaliana. Plant J 79(2):348–359. https://doi.org/10.1111/tpj.12554
Feng Z, Zhang B, Ding W, Liu X, Yang DL, Wei P (2013) Efficient genome editing in plants using a CRISPR/Cas system. Cell Res 23(10):1229–1232. https://doi.org/10.1038/cr.2013.114 PMID: 23958582
Nagappan J, Rozana R, Ting NC, Ooi LC-L, Low ETL, Singh R (2013) Exploiting synteny between oil palm and rice to find markers more closely linked to selected trait. J Oil Palm Res 25:180–187
Kalyana Babu B, Mary Rani KL, Sarika Sahu RK, Mathur Naveen Kumar P, Ravichandran G, Anitha P, Bhagya HP (2019) Development and validation of whole genome-wide and genic microsatellite markers in oil palm (Elaeis guineensis Jacq.): first microsatellite database (OpSatdb). Sci Rep 9:1899
Parveez GKA, Rasid OA, Masani MYA, Sambanthamurthi R (2015) Biotechnology of oil palm: strategies towards manipulation of lipid content and composition. Plant Cell Rep 34(4):533–543. https://doi.org/10.1007/s00299-014-1722-4
Masura SS, Tahir NI, Rasid OA, Ramli US, Othman A, Masani MYA, Parveez GKA, Kushairi A (2017) Post-genomic technologies for the advancement of oil palm research. J Oil Palm Res 29(4):469–486
Masani MYA, Izawati AMD, Rasid OA, Parveez GKA (2018) Biotechnology of oil palm: current status of oil palm genetic transformation. Biocatal Agric Biotechnol 15:335–347. https://doi.org/10.1016/j.bcab.2018.07.008
Masli DIA, Parveez GKA, Yunus AMM (2009) Transformation of oil palm using Agrobacterium tumefaciens. J Oil Palm Res 21:643–652
Parveez GKA, Masri MM, Zainal A, Majid NA, Yunus AMM, Fadilah HH, Rasid O, Cheah SC (2000) Transgenic oil palm: production and projection. Biochem Soc T 28(6):969–972. https://doi.org/10.1042/bst0280969
Masani MYA, Noll GA, Parveez GKA, Sambanthamurthi R, Prüfer D (2014) Efficient transformation of oil palm protoplasts by PEG-mediated transfection and DNA microinjection. PLoS One 9(5):e96831. https://doi.org/10.1371/journal.pone.0096831
Wang Y, Cheng X, Shan Q, Zhang Y, Liu J, Gao C (2014) Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew. Nat Biotechnol 32(9):947–951. https://doi.org/10.1038/nbt.2969 PMID: 25038773
Lee K, Zhang Y, Kleinstiver BP, Guo JA, Aryee MJ, Miller J, Wang K (2018) Activities and specificities of CRISPR/Cas9 and Cas12a nucleases for targeted mutagenesis in maize. Plant Biotechnol J 17:362–372
Jiang WZ, Henry LM, Lynagh PG, Comai L, Cahoon EB, Weeks DP (2017) Significant enhancement of fatty acid composition in seeds of the allohexaploid, Camelina sativa, using CRISPR/Cas9 gene editing. Plant Biotechnol J 15(5):648–657. https://doi.org/10.1111/pbi.12663
Morineau C, Bellec Y, Tellier F, Gissot L, Kelemen Z, Nogue F, Faure JD (2017) Selective gene dosage by CRISPR-Cas9 genome editing in hexaploid Camelina sativa. Plant Biotechnol J 15(6):729–739. https://doi.org/10.1111/pbi.12671
Amin N, Ahmad N, Nan W, Xiumin P, Tong M, Yeyao D, Xiaoxue B, Nan W, Sharif R, Piwu W (2019) CRISPR-Cas9 mediated targeted disruption of FAD2–2 microsomal omega-6 desaturase in soybean (Glycine max.L). BMC Biotechnol 19(1):9. https://doi.org/10.1186/s12896-019-0501-2
Wu N, Lu Q, Wang P, Zhang Q, Zhang J, Qu J, Wang N (2020) Construction and analysis of GmFAD2-1A and GmFAD2-2A soybean fatty acid Desaturase mutants based on CRISPR/Cas9 technology. Int J Mol Sci 21(3):1104. https://doi.org/10.3390/ijms21031104
Ayako O, Takumi O, Chie K, Kanako K, Mizue I, Jun I, Nobuya K (2018) CRISPR/Cas9-mediated genome editing of the fatty acid desaturase 2 gene in Brassica napus. Plant Physiol Biochem 131:63–69
Yuan M, Zhu J, Gong L, He L, Lee C, Han S, Chen C, He G (2019) Mutagenesis of FAD2 genes in peanut with CRISPR/Cas9 based gene editing. BMC Biotechnol 19(1):24. https://doi.org/10.1186/s12896-019-0516-8
Tian Y, Chen K, Li X, Zheng Y, Chen F (2020) Design of high-oleic tobacco (Nicotiana tabacum L.) seed oil by CRISPR-Cas9-mediated knockout of NtFAD2-2. BMC Plant Biol 20:233
Doudna JA, Charpentier E (2014) Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science 346(6213):1258096. https://doi.org/10.1126/science.1258096
Acknowledgements
The authors thank the management of MPOB for permission to publish this paper. The authors would like to thank all Transgenic Technology Group, Malaysian Palm Oil Board (MPOB), Malaysia, especially Nurul Huda Ideris, for their assistance. We gratefully acknowledge Prof. Kaijun Zhao, Prof. Chunlian Wang, Dr. Zhiyuan Ji, Yongchao Tang, Zheng Wei, and Hongda Sun of Institute of Crop Sciences, Chinese Academy of Agricultural Sciences, Beijing, China, for their generous support, coaching, and companionship during the training. We also thank Prof. Yaoguang Liu (South China Agricultural University, China) for kindly giving us permission to use his binary vector pYLCRISPR/Cas9 and the sgRNA plasmids.
Funding
The MPOB board fully funded this research approved project “Establishment of transgenic oil palm with high added value for commercial exploitation” (T0003050000-RB01-J).
Author information
Authors and Affiliations
Contributions
BB performed the experiments and drafted the manuscript. ROA and PGKA revised the manuscript. MMYA initiated and guided the project and critically revised the manuscript. The authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
This article does not contain any studies related to humans or animals.
Consent for publication
Not applicable
Competing interests
The authors declare that they have no known competing financial interests or personal relationships that could have influenced the work reported in this article.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1: Supplementary Table 1. List of primers used in this study. Supplementary Fig. 1
Regeneration of rice (O. sativa L.) cultivar Nipponbare used for CRISPR/Cas9 sgRNA transformation. Rice seedling growth stages, seed (A & B) germination (C), sprout (D) and seedling (E & F). Calli regenerated from mature seed used for biolistic mediated transformation, before (G) and after (H) bombardment. Supplementary Fig. 2 Construction of pYLCRISPRCas9PubiH-OsFAD. (a) pYLCRISPR/Cas9PubiH vector backbone digested with BsaI for insertion of sgRNAs. (b) First-round amplification of each sgRNA expression cassette, bands at ~600 bp (Lane 1: pYLsgRNAOsU6a) and ~500 bp (Lane 2: pYLsgRNAOsU6b) were amplified using the U-F/sgRNA reverse primers. (c) The bands at ~150 bp (Lanes 1 and 2) were amplified using the sgRNA forward /GR-R primers. PCR amplification of two sgRNAs expression cassettes, (d) U6aOsFAD2T1 and (e) U6bOsFAD2T2. (f) Verification of the CRISPR/Cas9 positive clones with Asc1 digestion. Lane M in (a), (b), (d), (e) and (f ) is Trans2K® Plus II DNA Marker (TransGen Biotech, China). Lane M in (c) is 100 bp Plus DNA ladder (TransGen Biotech, China). (e) The overall cloning strategy for construction of pYLCRISPRCas9Pubi-H:OsFAD2 using overlapping PCR to combine two sgRNAs expression cassettes and then cloned into pYLCRISPR/Cas9PubiH vector via gibson assembly cloning method. LB: left border, HPT: gene for hygromycin phosphotransferase, 2xP35S: Double cauliflower mosaic virus 35S promoter, Pubi: maize ubiquitin Ubi1 promoter, Cas9p: CRISPR associated protein 9, U6a: rice (O. sativa) U6a (OsU6a) promoter, sgRNA1: OsFAD2-T1, U6b: rice (O. sativa) U6b (OsU6b) promoter, sgRNA2: OsFAD2-T2, RB: right border). Supplementary Fig. 3 Recombinant screening analysis of pYLCRISPRCas9PubiH-OsFAD vector. (A) Gel analysis of recombinants via colony PCR using OsFAD2U6bT2R and OsFAD2U6aT1F primers. Lanes 1 to 11 are colonies purified and cloned. A red arrow with a size of 500 bp indicates PCR products. Lane M is Trans2K® Plus II DNA Marker (TransGen, Beijing, China). (B) The output from Sanger sequencing analysis. Underlined sequences consist of OsFAD2-T1 and OsFAD2-T2 sgRNAs. Orange and red letters indicated the OsU6a and OsU6b promoters, respectively. Supplementary Fig. 4 Molecular analysis of T0 transgenic rice calli. (A) The PCR products of 767 bp and 474 bp amplified using FAD2-F2 and FAD2-R primers. Lane M is Trans2K® Plus II DNA Marker (TransGen Biotech, China). (B) Sequence alignment for detection of mutations at target sites of OsFAD2-T1 and OsFAD2-T2. Sequence analysis of the edited line indicated in the light red box and dotted lines (….) represents deletions. The red and orange boxes are the target sequences, and the PAM sequence is shown in blue underlined nucleotides. (C) A schematic representation of the residue of target 1 (OsFAD2-T1) in the red box and the residue of target 2 (OsFAD2-T2) in the orange box. Supplementary Fig. 5 (A) Sequencing result decoded homozygous mutations in both alleles. (B) Representative chromatograms of PCR product from homozygous mutant alleles with expected (i) 302 nucleotides deletion for amplicon 474 bp and (ii) 1 nucleotide insertion for amplicon 776 bp in the T0 rice.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Bahariah, B., Masani, M.Y.A., Rasid, O.A. et al. Multiplex CRISPR/Cas9-mediated genome editing of the FAD2 gene in rice: a model genome editing system for oil palm. J Genet Eng Biotechnol 19, 86 (2021). https://doi.org/10.1186/s43141-021-00185-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s43141-021-00185-4